Discriminative Random Walks with Restart (DRaWR)
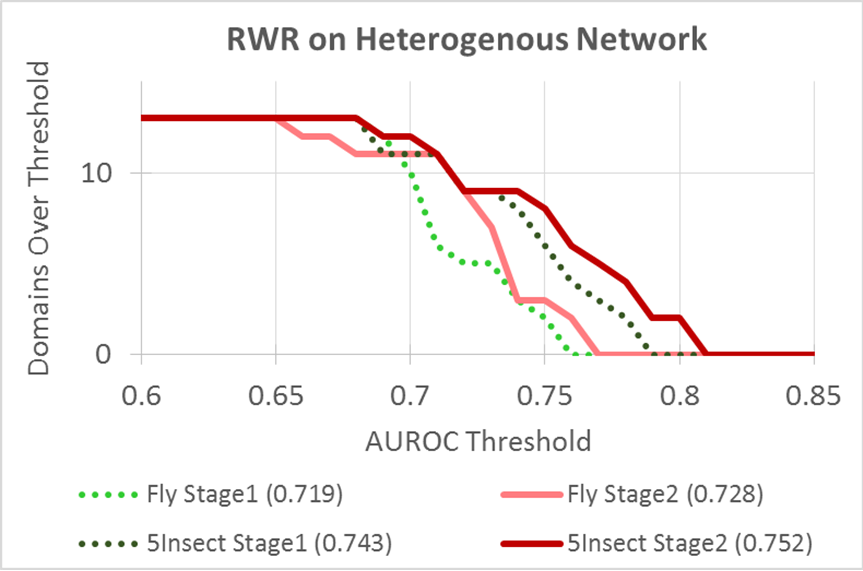
DRaWR is a network-based method for ranking genes or properties related to a given gene set that is designed to operate on large, heterogeneous networks of biological information. Our method involves a random walk with restarts, performed on an initial network with multiple node and edge types that preserves more of the original, specific property information than homogeneous networks typically utilized by related methods. In this first stage of our algorithm, we find the property nodes that are the most relevant to the given gene set and extract a subnetwork of the original network, comprising only the relevant properties. We then rerank genes by their similarity to the given gene set, based on a second random walk with restarts, performed on the relavent subnetwork. We demonstrate the effectiveness of this algorithm for ranking genes related to Drosophila embryonic development and aggressive responses in the brains of social animals.
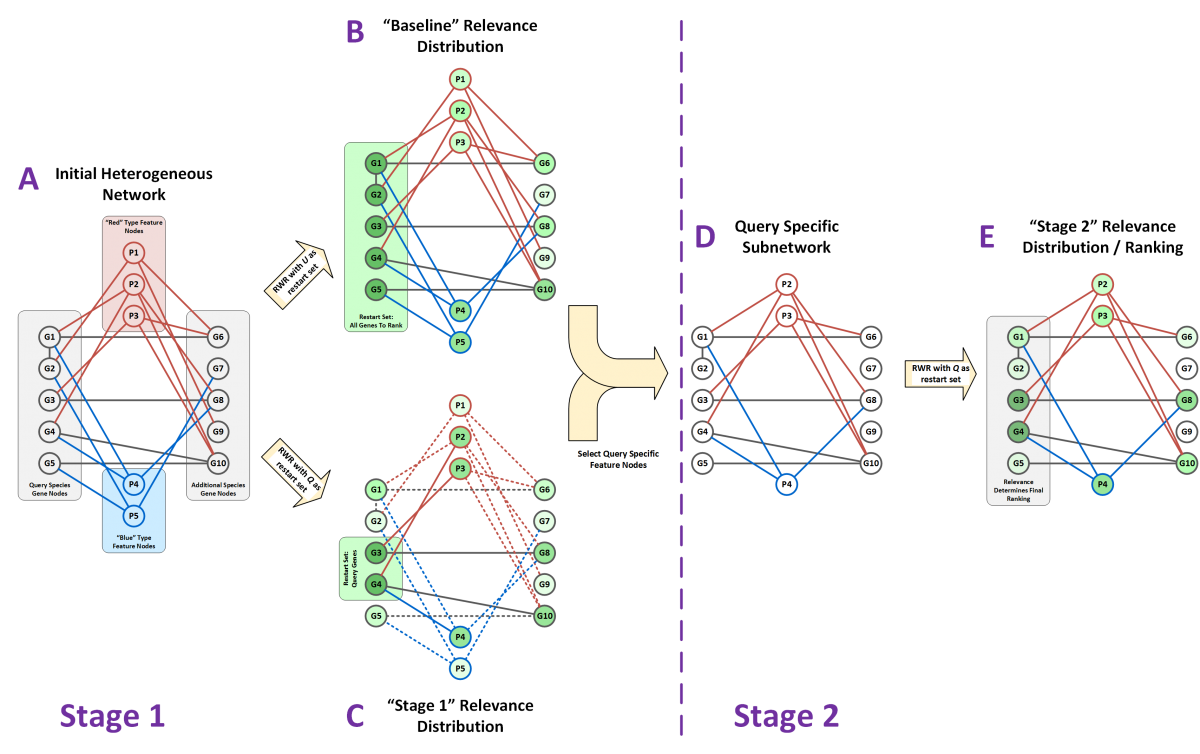
DRaWR is used in this example to rank all genes from Species 1 (G1,G2,…,G5) based on their relatedness to the query set of genes (G3,G4). A) A network with gene nodes (gray) from two species is connected by protein sequence homology edges (black) and through two types of property nodes (red and blue). B) A random walk with restart is run with all Species 1 genes as the restart set to understand the baseline relevance of all nodes in the network. C) A random walk with restart is run with the query nodes as the restart set. The difference between the query specific relevance and the baseline relevance is used to select a subset of query specific feature nodes (P2, P3, and P4 in this example). D) A subnetwork of the original is made containing only feature nodes that are query specific. E) A final random walk is run on Stage 2 subnetwork with the query set as the restart set once again. The relevance scores from this final random walk are used to provide the final ranking on Species 1 genes for similarity to the query set.